Authors: Ryan Cook, Ryan Barnhart, and Sudipta Majumdar
Institution information: Department of Chemistry, Indiana University of Pennsylvania, 975 Oakland Ave, Indiana, PA 15705
ABSTRACT
Selective pressure generated by the misuse of antibiotics in medicine and agriculture has resulted in antimicrobial resistance and the subsequent reemergence of several pathogenic bacteria, including Mycobacterium tuberculosis (MT), the etiological agent of tuberculosis (TB) disease. Increased resistance of MT to previously-effective antibiotics is associated with greater incidence rates and burden of TB disease. Novel approaches to infection mitigation must be explored if we are to attenuate the destructive force of this pathogen. Alanine racemase (Alr), a ubiquitous bacterial enzyme required for peptidoglycan biosynthesis, is a known target for antibacterial drug development that has not been fully exploited. It is critical to assign pH optima for the proper characterization and utilization of Alr as a drug target. Here, we report a maximum k_cat of 1.63 ± 0.03 s^-1 at pH 10 and a lowest K_m of 0.700 ± 0.053 mM at pH 9. A pH optimum for k_cat/K_m at pH 9.1 for was found for Alr from the human pathogen M. tuberculosis. These results will hopefully facilitate future investigations of this potentially potent antibacterial drug target.
INTRODUCTION
Tuberculosis (TB), the pathological consequence of occult colonization in the lung of the gram-positive bacterium Mycobacterium tuberculosis (MT), is resurfacing as a major infectious disease around the globe especially in impoverished developing countries. According to the Centers for Disease Control and Preventions, approximately one-third of the world population is currently infected with TB, and the appearance of multidrug resistant (MDR) and extremely drug resistant (XDR) strains of MT have contributed to the perpetuation of TB disease as an ongoing world health crisis (Sakamoto, 2012). An estimated 20% of MT isolates are resistant to at least one major drug. Approximately 5% of TB patients globally are infected with either MDR or XDR strains, and of the MDR infected individuals an estimated 10% are infected with either XDR MT or MT resistant to additional drugs beyond XDR MT (totally drug resistant; Dheda et al., 2017). Therefore, there is a continuing need for new anti-tuberculosis targets and drugs.
One well-known target whose potential has yet to be fully explored is the pyridoxal phosphate (PLP)-dependent bacterial amino isomerase, alanine racemase. Alanine racemase (Alr; EC 5.1.1.1) isomerizes L-alanine to D-alanine for cell wall biosynthesis (Lambert and Neuhaus, 1972). It is well established that in bacterial systems insufficient environmental D-alanine results in an endogenous demand satisfied by Alr (Milligan et al., 2007). Because this protein has such a profound effect on the survivability of bacteria and is ubiquitous in bacterial systems, yet absent in higher eukaryotes, it has been recognized as an attractive target for mitigating bacterial infection (Milligan et al., 2007).
Recent studies have shown that the Alr mutant strains of M. tuberculosis require D-alanine for growth, establishing the importance of Alr for the survival of this bacterium (Awasthy et al., 2012). In order to efficiently study and ultimately develop novel inhibitory agents a sufficient structural and functional profile of MT_Alr must be established. The structural characteristics of MT-Alr have already been documented (LeMagueres et al., 2005). In this paper, we report the cloning, overexpression, purification, and functional characterization of MT_Alr. These results will facilitate the design of novel anti-tuberculosis agents.
MATERIALS AND METHODS
All reagents and chemicals used as buffers and substrates of the highest available chemical grade were purchased from Sigma-Aldrich, Fisher, Acros, or Alfa Aesar. The buffers were prepared by mixing 0.1 M sodium phosphate monobasic, 0.1 M sodium phosphate dibasic, 0.1 M sodium carbonate, and 0.1 M sodium bicarbonate to pH values of 7.0, 7.5, 8.0, 9.0, 9.5, 10.0, and 10.5. The UV-Vis spectrophotometric data were collected using Multiskan GO (Fisher Scientific, Waltham, MA). Protein purifications were conducted using BioLogic DuoFlow (Bio-Rad, Hercules, CA). The genomic DNA for MT (ATCC 25177) was purchased from American Type Culture Collection (Rockville, MD). L-alanine dehydrogenase was cloned, expressed and purified in the lab from Streptomyces coelicolor (data not shown).
Plasmid construction
Figure 1. Recombinant pET28B-MT_Alr plasmid map. MT_Alr gene indicated by arrow.
The genes encoding the Alr from MT was amplified via PCR from their corresponding genomic DNA with the primers (Integrated DNA Technologies, Coralville, IA) listed in Table 1. The PCR mixture (50 μL) contained 1 μL of 100 ng/μL template genomic DNA, 25 μL of Phusion master mix (2X), 1.5 μL of dimethyl sulfoxide 2.5 μL of 10 μM forward primer, 2.5 μL of 10 μM reverse primer, 0.5 μL of Phusion High Fidelity (2 U/μL) enzyme (Thermo Scientific, Waltham, MA), and 17 μL of ddH2O. The PCRs were performed in a T100 Thermal Cycler (Bio-Rad, Hercules, CA) with the following parameters: 98°C for 30 s followed by 30 cycles of 98°C for 10 s, 60°C for 20 s, and 72°C for 30 s, with a final extension time of 7 min at 72°C. The PCR products were purified by gel extraction (Qiagen, Venlo, Netherlands) following the manufacturer’s protocol. The amplified DNA product was digested with the BamHI and XhoI restriction enzymes (New England Biolabs, Ipswich, MA) and ligated into the pET28b expression vector (Novagen, Madison, WI) cut with the same enzymes, creating the recombinant plasmid pET28b-MT_Alr (Figure 1).
Enzyme expression and purification
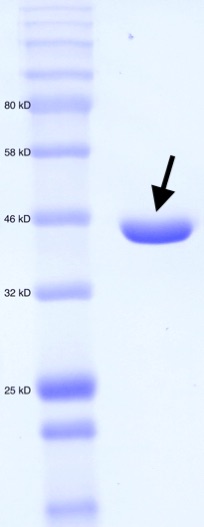
Figure 2. SDS-PAGE for purified MT_Alr. Molecular weight standard (left). Purified MT_Alr (indicated by arrow).
The enzyme MT_Alr cloned with an N-terminal His-tag, was expressed in E. coli BL21 (DE3) cells at 20˚C. A bacterial culture for a typical large-scale purification utilized 2X 1L of TB medium shaken at 37˚C until the OD600 reached 0.6 at which point expression was induced to the final concentration of 1 mM IPTG. The culture was incubated for a further 12 hr at 37˚C with shaking. Cells were subsequently harvested by centrifugation. The cell pellets were dissolved in 35 mL of cold buffer (20 mM 2-Amino-2-(hydroxymethyl)propane-1,3-diol (Tris), 100 mM NaCl, pH 8.0). Cells were lysed by sonication and the lysate was cleared by centrifugation. The supernatant was applied to a 5 mL Bio-Scale Mini Nuvia IMAC column (Bio-Rad, Hercules, CA) and eluted with a linear gradient (100 mL) of 1M imidazole buffered with 20 mM Tris and 100 mM NaCl (pH 8.0). Fractions containing pure (>95%) Alr were collected and dialyzed against 20 mM Tris-HCl, 100 mM NaCl (pH 8.0) and then stored at -80˚C. The purity of the MT_Alr fractions was verified by SDS-PAGE (Figure 2).
Determination of enzyme activity at different pH values
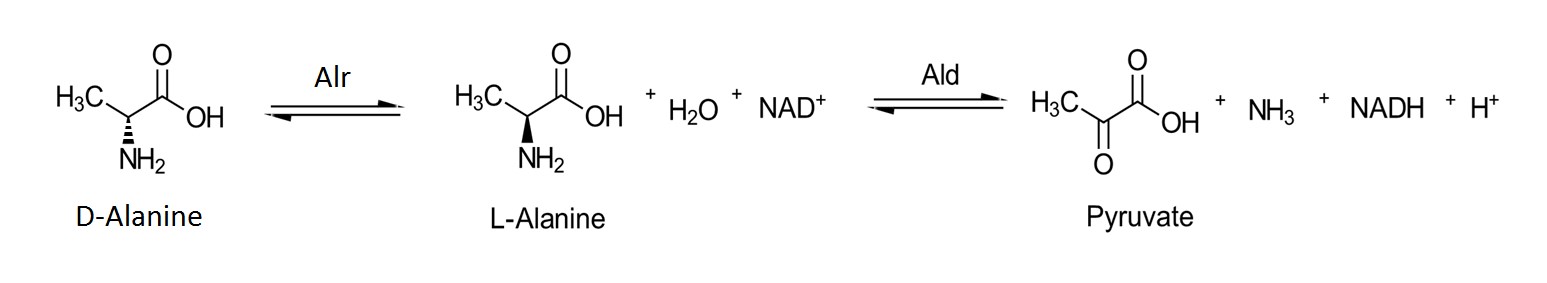
The MT_Alr activities at different pH values ranging from 7 – 10.5 were determined at 30˚C using D-alanine as the substrate. The Alr activity was measured in the D- to L-alanine direction by monitoring the production of NADH (ε_340 = 6.22 mM-1cm-1) at 340 nm as the L-alanine was converted to pyruvate and ammonia by L-alanine dehydrogenase (Figure 3). The reaction mixture (250 µL) contained 0.01 – 50 mM D-alanine (dissolved in buffer), NAD (906 μM), L-alanine dehydrogenase (2.76 µM), and Alr (0.6 µM) with appropriate buffer. The steady-state kinetic constants were determined by fitting the kinetic data to Michaelis-Menten equation for each pH using GraphPad Prism7 (GraphPad Software, Inc., La Jolla, CA).
Figure 3. Isomerization of D-alanine to L-alanine in the presence of Alr followed by the NAD+ dependent oxidation of L-alanine to pyruvate in the presence of alanine dehydrogenase.
RESULTS
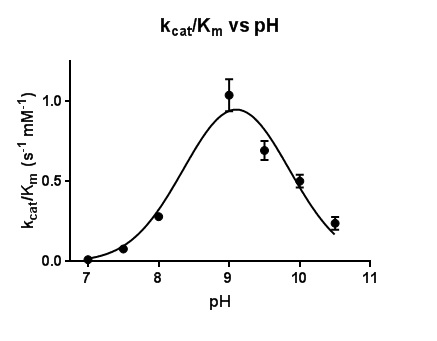
The steady-state kinetic parameters are reported in Table 2. Under the experimental conditions, the k_cat values increased as pH increased, reaching the maximum k_cat value, 1.63 ± 0.03 s^-1, at pH 10. On the other hand, K_m values decreased with increasing pH, reaching a minimum K_m value, 0.70 ± 0.05 mM, at pH 9. Beginning at pH 9, K_m values increased with increasing pH. The optimum k_cat/K_m value was found to be at pH 9.1.
DISCUSSION
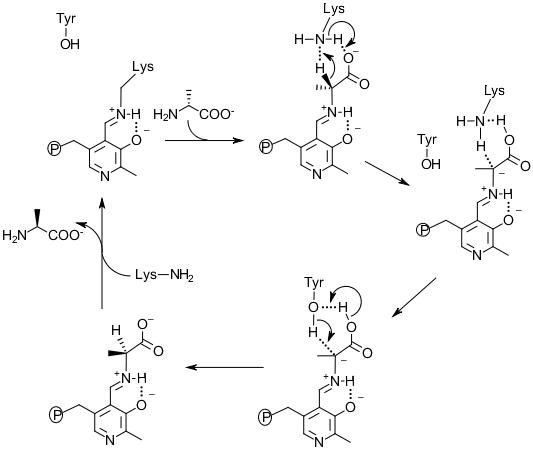
Figure 4. The mechanism of Alr catalysis for D-alanine to L-alanine direction.
Structural, site-directed mutagenesis and kinetic studies of Alr from Bacillus Stearothermophilus (Shaw et al., 1997; Stamper et al., 1998; Watanabe et al., 1999; Watanabe et al., 1999) support the description of the two base mechanism involving Lys39 and Tyr265’ residues (Watanabe et al., 2002). The MT_Alr crystal structure supports the likelihood of this proposed mechanism with conserved Lys42 and Tyr271’ residues at the active site (LeMagueres et al., 2005). This mechanism (Figure 4) proposes that, in the D-alanine to L-alanine direction, the substrate enters the active site and forms an external aldimine bond with the PLP cofactor. The α-hydrogen is then removed by the Lys42 residue. The Tyr271’ donates a proton to the opposite side of the α-carbon of the intermediate resulting in L-alanine formation. The likelihood of such a mechanism depends on the presence of a mediator transferring protons between the two catalytic residues, Lys42 and Tyr271’, in order to reset these residues to the requisite protonation state, i.e. Lys42 deprotonated and Tyr271’ protonated. Without such a mediator, the enzyme reaction will stop after a single turnover. The carboxylate group of the substrate alanine is proposed to serve as the mediator transferring protons between two catalytic residues during the catalytic cycles (Sun and Toney, 1998).
Figure 5. Plot of the catalytic efficiency k_cat/K_m for the reaction of MT_Alr with D-alanine. The line represents the best fits of the data using nonlinear regression curve fitting.
The dependence of k_cat and K_m on pH in this work is reflected in the bell-shaped curve of the activity versus pH plot, which reaches a maximum value at pH 9.1 (Figure 5). Two pK_a values, 7.0 and 10.5, are represented in the findings reported here. The deprotonation of the hydroxyl group of the Tyr271’ is most likely responsible for the lower pKa value (7.0) on the pH profile. Extensive hydrogen bonding network in the active site, specifically, the close proximity of the positively-charged residues, Arg140 and His172, to the catalytic Tyr271’ (LeMagueres et al., 2005), explains the lowering of the pKa value from 10.5 to 7.0 for the Tyr271’ hydroxyl group under catalytic conditions. The higher pKa value of 10.5 on the pH profile is assigned to the amine side chain (pKa 10.5) of the Lys42 under catalytic conditions. Therefore, the bell-shaped profile with pKa 7.0 and 10.5 appears to reflect the reaction of the protonated of Tyr271’ donating a proton to the intermediate and the deprotonated Lys42 accepting the α-hydrogen from the bound substrate.
This field of research will remain relevant as pathogenic bacteria continue to acquire resistance to the drug therapies currently employed. This research is a lateral step, providing a better position from which a viable drug target, Alr, can be explored. The intent of this work is to provide the information necessary to explore the effects of compounds that possess Alr inhibitory qualities so that the pathogenic effects of M. tuberculosis may be mitigated and the worldwide burden associated with this infectious agent may be lessened.
ACKNOWLEDGMENTS
We thank Indiana University of Pennsylvania for financial support.
REFERENCES
Awasthy D, Bharath S, Subbulakshmi V, and Sharma U. Alanine racemase mutants of Mycobacterium tuberculosis require D-alanine for growth and are defective for survival in macrophages and mice. Microbiology (Reading, England; 2012), 158(Pt 2), 319-27.
Dheda K, Gumbo T, Maartens G, Dooley KE, McNerney R, Murray M, …Warren RM. The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respiratory Medicine (2017), 5:4, 291-360.
Lambert MP and Neuhaus FC. Factors affecting the level of alanine racemase in Escherichia coli. Journal of Bacteriology (1972), 109:3, 1156-61.
LeMagueres P, Im H, Ebalunode J, Strych U, Benedik MJ, Briggs JM, … Krause, KL. The 1.9 A crystal structure of alanine racemase from Mycobacterium tuberculosis contains a conserved entryway into the active site. Biochemistry (2005), 44:5, 1471-81.
Milligan DL, Tran SL, Strych U, Cook GM, and Krause KL. The alanine racemase of Mycobacterium smegmatis is essential for growth in the absence of D-alanine. Journal of Bacteriology (2007), 189:22, 8381-6.
Sakamoto K. The pathology of Mycobacterium tuberculosis infection. Veterinary Pathology (2012), 49:3, 423-39.
Shaw JP, Petsko GA, and Ringe D. Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9-A resolution. Biochemistry (1997), 36:6, 1329-42.
Stamper GF, Morollo AA, and Ringe D. Reaction of alanine racemase with 1-aminoethylphosphonic acid forms a stable external aldimine. Biochemistry (1998), 37:29, 10438-45.
Sun S and Toney MD. Evidence for a two-base mechanism involving tyrosine-265 from arginine-219 mutants of alanine racemase. Biochemistry (1998), 38:13, 4058-65.
Watanabe A, Kurokawa Y, Yoshimura T, and Esaki N. Role of tyrosine 265 of alanine racemase from Bacillus stearothermophilus. Journal of Biochemistry (1999), 125:6, 987-90.
Watanabe A, Kurokawa Y, Yoshimura T, Kurihara T, Soda K, and Esaki N. Role of lysine 39 of alanine racemase from Bacillus stearothermophilus that binds pyridoxal 5’-phosphate. Chemical rescue studies of Lys39 --> Ala mutant. Journal of Biological Chemistry (1999), 274:7, 4189-94.
Watanabe A, Yoshimura T, Mikami B, Hayashi H, Kagamiyama H, and Esaki N. Reaction mechanism of alanine racemase from Bacillus stearothermophilus: x-ray crystallographic studies of the enzyme bound with N-(5’-phosphopyridoxyl)alanine. Journal of Biological Chemistry (2002), 277:21, 19166-72.