Author: Eleanor Sheekey
Proteins are often intuitively named after their function, and condensin is no exception. It condenses DNA inside the nucleus of a cell into X-shaped chromosomes for cell division. The name, however, provides no insight into how this feat is achieved. In fact, the molecular mechanism of how condensin organizes DNA inside the nucleus has previously only been hypothesized to involve extruding DNA into large loops. However, a recent Science paper by Ganji et al.,(1)from the Kavli Institute of Delft University and EMBL Heidelberg,provides real-time imaging that shows condensin in action.
DNA is packaged inside the nucleus in a structure known as chromatin. DNA packaging is not a simple taskand is proposed to occur by the successive supercoiling and looping of chromatin. This looping allows the cell to create chromosomes, the highest orderof chromatin compaction, which are necessary for cellular division.Although it is currently unclear which exact higher-order structure is required to create chromosomes(2), it is believed that looping plays a key part in the organization. Since looping can bring two regions of DNA distant in sequence into close proximity to interact, the mechanism and regulation of this process have been of great interest, particularly for their influence on cell division.
Condensin is a member of the structural maintenance of chromosomes (SMC) family that includes cohesin, the protein that is involved in sister chromatid cohesion during cell division. The proteins are characterized by their ability to form ring-like structures that are wide enough to encircle DNA strands. Condensin has been linked with chromatin condensation but its mechanism had not been fully elucidated. However, the Ganjigroup’s latest research now providesconvincing evidence supporting the active role played by condensin in DNA loop formation. This builds upon work published last year in Science that showed condensin to be a motor that translocates along DNA,(3)providing a clearer picture of DNA compaction.
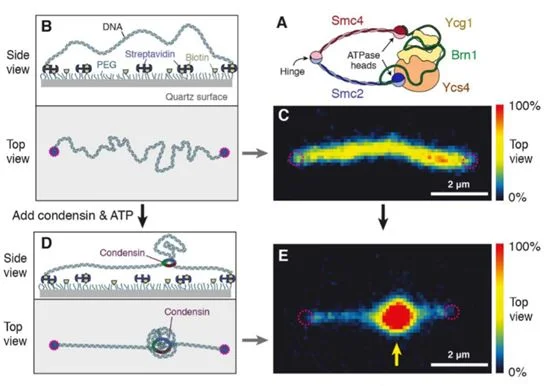
By tethering a 48.5 kilobase pair segment of DNA to a surface and staining with Sytox Orange (SxO), the team couldvisualize the structure and density of DNA in real time.Condensin was added, allowing the protein to bind DNA, and successive images were takenover time to observe changes in movement shown by the fluorescent staining (Figure 1).DNA extrusion was dependent on the presence of ATP to provide energy for condensin to function.The team noticed a gradual increase in fluorescent intensity, which confirmed that condensin creates DNA loops by slowly extruding DNA, instead of grabbing two sections of DNA and bringing them together.
Figure 1: Experimental set-up to visualizecondensin-mediated DNA extrusion (Figure taken from reference (1).
To test if condensin was reeling out DNA from both or only one side of the strand, Ganji’s group studied the change in DNA length on either side of where the loop was forming. Interestingly, while DNA length decreased on one side, the other end showed no change throughout the experiment. This strongly supported an asymmetric mechanism of loop extrusion. For this to work, condensin must remain firmly attached to the DNA strand in which it is extruding.This contrasts to what was previously hypothesized from computer loop-extrusion models and, thus, this finding will greatly contribute to improved DNA compaction reconstructions.
Condensin and other SMC proteins play important roles in organizing the 3D architecture of the genome throughout all domains of life. Genome organization has functional relevance to both the execution of cell division and defining gene expression. Since errors in genome organization can ultimately result in cancer and other genetic diseases,knowing the mechanism of condensin will help understand the cause of these defects.For example,mutations in SMC proteins have been associated with Cornelia de Lange syndrome, a genetic disorder that causes short stature and intellectual disabilities.Although these studies have been performed in artificial conditions, they provide an explanation for the long-held mystery of the nature of DNA compaction.However, further experiments in more physiological conditions are needed to validate these results.
References
(1) M. Ganjiet al., Science 10.1126/science.aar7831 (2018)
(2) Ou et al., Science 357, 370 (2017)
(3) T. Terakawa et al., Science 10.1126/science.aan6516 (2017)