Authors: Stephanie N. Stotts , Olivia D. Nigro, Tonya L. Fowler , Roger S. Fujioka, and Grieg F. Steward
Institution: Baker University
Date: April 2005
Abstract
Staphylococcus aureus is often found passively colonizing human skin and nasal passages. As an opportunistic pathogen, the bacterium can also cause infections that vary widely in their severity and in their susceptibility to antibiotic treatment. This variability is caused by differences in gene content among strains. One source of exposure to novel, potentially more virulent strains of S. aureus is recreational swimming areas, since S. aureus is readily shed from humans into water. The standard measure of S. aureus contamination in recreational waters is the concentration of colony forming units, but this method does not discriminate among strains of differing virulence. As a first step toward characterizing the diverse strains of S. aureus to which beachgoers may be exposed, sixteen strains were isolated from seawater around Oahu and tested for the presence of eight genes associated with either virulence (tst, cna, ica, hlg[B and C], sea, and sdrE) or antibiotic resistance (mecA, femA) using the polymerase chain reaction and gene-specific primers. The maximum number of these genes observed in any single isolate was six and the minimum was two with a total of nine different gene combinations observed among the sixteen isolates. The results show that the virulence gene content of S. aureus strains isolated from coastal waters varies widely. Further characterization of the diversity of S. aureus in the environment could, therefore, contribute to a better understanding of the epidemiology of community-acquired S. aureus infections and lead to improved measures of the exposure risk for swimmers in recreational waters.
Introduction
Staphylococcus aureus is an opportunistic human pathogen capable of causing a wide variety of diseases including septicemia, pneumonia, wound sepsis, septic arthritis, osteomyelitis, food poisoning, and toxic shock syndrome (Boyd and Brüssow, 2002; Moore and Lindsay, 2001). Most people are passive carriers of S. aureus, which is often found colonizing the nasal passages (Kluytmans et al., 1997), and in some cases, an infection may arise from self-inoculation of a wound. S. aureus infections have been a major problem in hospitals for decades, but the incidence of community-acquired infections has also been increasing (Chambers, 2001).
The type as well as severity of an S. aureus infection and its response to antibiotic treatment are dictated by the specific suite of virulence- and antibiotic resistance-associated genes carried by the strain of S. aureus causing the infection (Peacock et al., 2002). There are over 40 virulence-associated genes identified among various strains of S. aureus, many of which are encoded by mobile genetic elements. Until recently, methicillin was one of the few remaining antibiotics effective against S. aureus, but the emergence and spread of methicillin-resistant S. aureus (MRSA) has severely decreased the usefulness of this antibiotic. While this problem was initially confined to clinical settings (Chambers, 2001), the frequency of community-acquired MRSA is becoming more common and some of these community-acquired strains are particularly virulent (Baba et al., 2002).
A correlation between seawater exposure and S. aureus infection rates (Charoenca and Fujioka, 1995) suggests that the oceans may be one pathway by which community-acquired infections are transmitted. S. aureus is readily shed from people while swimming or engaging in other water-related activities (Charoenca and Fujioka, 1993) and high counts of S. aureus in recreational waters is often considered a risk factor for contracting many diseases that affect areas such as the skin, eyes, and ears (Gabutti et al., 2000). Although S. aureus is routinely found in the waters surrounding the island of Oahu in the state of Hawaii (Charoenca and Fujioka, 1995; Charoenca and Fujioka, 1993), nothing is known yet about the distribution of virulence genes among the various strains recovered from environmental sources. Most investigations of genetic diversity have focused on isolates collected directly from healthy or diseased humans (Diep et al., 2004; Monk et al., 2004; Nashev et al., 2004). To better evaluate risk, it would be helpful to determine not only total counts of S. aureus in recreational waters, but also the number and type of virulence-associated genes that they carry.
Recent studies suggest that adaptations to the external environment (Wilson and Salyers, 2003) and lateral gene transfer among strains (Boyd and Brüssow, 2002; Fitzgerald et al., 2001) may play important roles in the evolution of pathogens. Knowledge of the genetic diversity of S. aureus in the environment could, therefore, shed light on the emergence and spread of novel pathogenic strains. In this preliminary study, S. aureus strains were isolated from several beaches around Oahu and from a canal in downtown Waikiki for partial genetic characterization. DNA from each isolate was then screened by the polymerase chain reaction (PCR) for the presence or absence of six virulence-associated genes and two genes associated with methicillin resistance.
Methods & Materials
Sample Collection
Water samples were collected by hand in acid-washed polycarbonate, or autoclaved polypropylene, containers from Kailua Beach on the eastern shore of Oahu, from Kuhio, Sans Souci, and Ala Moana beaches on the southern shore of Oahu, and from the Ala Wai canal, which runs inland along the northern edge of Waikiki. For sampling at beaches, the collector waded into knee- to waist-deep water and submerged the collection vessel in the vicinity of swimmers. The vessel was kept in front of the collector who moved forward through the water as the vessel filled. At the Ala Wai canal, a bottle was submerged from a boat dock without requiring contact of the collector with the water. Bottles were immediately capped and handled aseptically through the isolation process.
Isolation and Cultivation of S. aureus
Bacteria were collected from seawater samples by filtration onto sterile, 47 mm-diameter, 0.45 um pore-size mixed cellulose ester filters (Pall Corporation, East Hills, New York, or Millipore Corporation, Billerica, Massachusetts). The filters were overlaid onto the selective medium CHROMagarTM Staph aureus (CSA; CHROMagar Microbiology, Paris, France) or onto CSA supplemented with oxacillin (4µg ml-1), sodium azide (0.0035%), or sodium azide plus glycine (12 g l-1). Alternatively, filters were incubated in Nutrient Broth with oxacillin to enrich for S. aureus prior to platting on solid media as above. Putative S. aureus colonies were picked from the filters and purified by serially picking and streaking individual colonies on fresh CSA or tryptic soy agar plates. Isolates were screened for methicillin resistance by plating on oxacillin-containing media. Putative MRSA isolates were confirmed using the disk diffusion test and a PBP2' agglutination assay.
DNA Purification
Each purified S. aureus isolate was grown to stationary phase in tryptic soy broth. Cells in 3 ml of culture were then pelleted by centrifugation for 10 minutes at 10,000 g then resuspended in 25 µl of TER Buffer (10 mM Tris-Cl, 1mM EDTA, 0.1% SDS, 100 µg ml-1 RNase A). Resuspended cells were extracted using a potassium ethyl xanthogenate protocol (Tillet and Neilan 2000) combined with bead beating. XS buffer was prepared immediately before use by combining 0.1 g of potassium ethyl xanthogenate (Sigma-Aldrich, Corp., Saint Louis Missouri), 1 ml of 1 M Tris at pH 7.4, 0.4 ml of 0.5 M EDTA at pH 8, 1 ml of 10% SDS, 2 ml of 4 M ammonium acetate, and 5.6 ml of ultrapure water (Nanopure Diamond, Barnstead International, Dubuque, Iowa). XS buffer (750 µl) was added to the resuspended pellets and the mixture transferred to 2 ml-capacity, screw-cap microcentrifuge tubes containing 0.5 g of 0.1 mm zirconium beads (Biospec Products, Inc. Bartlesville, Oklahoma). Samples were shaken in a FastPrepTM instrument (Q-Biogene, Carlsbad, California) for 30 s at a setting of 6 m s-1. All samples were then incubated at 70 °C for 1 hour followed by brief vortex mixing and incubation on ice for 30 minutes. The samples were centrifuged at 14,000 rpm for 10 minutes at 4°C to pellet precipitated protein-SDS complexes and the supernatants containing the extracted DNA were transferred to fresh tubes.
Precipitating DNA
GlycoBlueTM (Ambion, Inc.) was added to each of the supernatants at a final concentration of 50 µg ml-1 to aid in precipitation. An equal volume of isopropanol was added, the samples were then mixed by several inversions of the tube, incubated for 10 min at room temperature, then centrifuged at 8,000 rpm for 30 minutes at 4° C. The supernatant was discarded and the pellet was washed by adding 1 ml of 70% ethanol followed by gentle mixing to rinse the entire inner surface of the tube. The pellet was centrifuged as before, but for only 10 min, then the supernatant was removed and the pellets were allowed to air dry completely. All pellets were resuspended in 50 µl of TE buffer (100mM of Tris-Cl and 10mM EDTA).
Quantifying Extracted DNA
DNA extracts were analyzed qualitatively by electrophoresis in a 0.8% agarose gel. DNA in the gel was stained with ethidium bromide (0.005%, w/v) and visualized on a UV transilluminator. Images of gels were captured with a digital gel documentation system (Gel Doc-It, U). DNA yields were also quantified by fluorometry using a DNA Quantification kit (Quant-iT DNA assay, Molecular Probes, Inc., Eugene Oregon) and a filter-based fluorometer (TD700, Turner Designs).
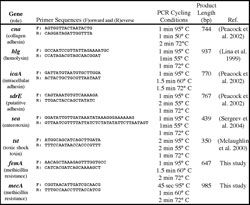
Table 1. Targeted genes and PCR conditions used for amplification. All PCR reactions began with a 95° C denaturation for 5 minutes and were terminated with 72° C extension for 7 minutes and a 4° C hold. Product length for can may vary by multiples of 561 bp as a result of sequence repeats of the B region. Due to sequence similarities, the primers for hlg will co-amplify hlgB and hlgC.
PCR amplification
Six virulence-associated genes (cna, tst, sdrE, sea, hlg[B and C], and ica) and two genes associated with antibiotic resistance (femA, mecA) were amplified from DNA extracts using the Expand High Fidelity PCR kit (Roche, Indianapolis, Indiana) using primers and thermal cycling protocols specific to each gene (Table1). The reaction contained 100 µmol primers and 0.5 µl template (6 to 31 ng) in a final reaction volume of 50 µl.
Cloning and Sequencing of Virulence Genes
A portion of each PCR product (2 µl) was ligated into the pCR 2.4-TOPO vector and transformed into a TOP10 chemically competent E. coli using the TOPO TA® Cloning kit (Invitrogen, Carlsbad California). Following transformation, 20 and 40 µl of the transformation mixture were spread onto LB agar plates which had been previously supplemented by spreading 100 µl of 2% x-gal (5-bromo-4-chloro-3-indolyl ß-D-galactopyranoside) per plate. Plates were incubated at 37°C overnight then two colonies of each type of clone were selected through blue/white screening and transferred to 2-ml tubes with air-permeable caps (Eppendorf® LidBAC, Brinkmann Instruments, Inc., Westbury, New York) containing1.5 ml of LB broth with kanamycin (Sigma St. Louis, Missouri). Tubes were incubated in a shaking incubator (VorTemp Microtube Incubator Shaker, Midwest Scientific, Valley Park, Missouri) at 37°C for 24 hours. After incubation, cells were pelleted by centrifugation for 10 minutes at 10,000 g at room temperature. Plasmid DNA was purified using a QIAprep Spin Miniprep kit per manufacturer's instructions (Qiagen). To verify that the selected clones contained an insert of the expected size, 2 µl of this solution was digested with the restriction enzyme EcoR1 and the product size was analyzed by gel electrophoresis on a 2% gel. To verify that the cloned products were the targeted virulence genes, one clone representing each gene was sequenced using CEQ Dye Terminator Cycle Sequencing with Quick Start kit (Beckman Coulter, Inc. Fullerton, California). The sequencing reactions were analyzed on a model CEQ 200XL sequencer (Beckman Coulter, Inc).
Table 2. The distribution of genes associated with antibiotic resistance and virulence among S. aureus strains isolated from the Ala Wai canal in Waikiki and various beaches around Oahu.
Results
Each of the genes could be successfully amplified by PCR from at least one of the isolates (Figure 1), but no isolate was positive for all eight of the genes. Overall, among the 16 isolates, nine different combinations of the targeted genes were found (Table 2). The most commonly occurring gene, sea, was found in all but one of the isolates tested. In contrast, cna and tst were found in only one isolate each. One genotype contained as few as two of the eight genes while eight genotypes contained six. Eight S. aureus isolates previously determined to be resistant to methicillin were shown to have the expected methicillin resistance-associated genes mecA and femA. However, five of eight isolates that were sensitive to methicillin by the disk diffusion test were also found to have both the mecA and femA genes.
Discussion
Although there are many known virulence-associated genes in S. aureus, this preliminary study focused on only a small subset. Some of these (cna, hlg, icaA, sdrE) were chosen because they were among those found to be more common among invasive isolates (Peacock et al., 2002). Assays for staphylococcal enterotoxin A (sea) and toxic shock toxin (tst) were included, because they can be transferred laterally among strains via bacteriophage infection (Ruzin et al., 2001; Betley and Mekalanos, 1985). The number of different combinations in which the eight selected genes were observed among the sixteen isolates indicates that genetic diversity of S. aureus in coastal waters of Oahu is high. There was no obvious correlation between the combination of genes found in an isolate and the location from which it was derived suggesting that the observed diversity is not restricted by geography. Few isolates from each site were analyzed, however, and some pattern to the distribution of genotypes may appear as additional isolates from more sites are analyzed.
Figure 1. Sizing of representative positive PCR amplification products for each targeted gene by agarose gel electrophoresis. First and last lanes are DNA size markers (M). Other lanes are PCR products from the genes as indicated, or negative controls marked with "-".
The virulence genes mecA and femA were examined because they are believed to be major contributors to methicillin resistance (Maidhof et al., 1991; Kuhl et al., 1978). While mecA and femA genes were found in all eight of the methicillin-resistant strains, they were also found in five methicillin-susceptible strains. In fact, the same combination of the eight tested genes was found in both methicillin-susceptible and a methicillin-resistant strains. The PCR-based assays used in this study only indicate presence or absence of a gene. The discrepancy between the resistance assays and gene content could therefore be due to differences in the expression of mecA and femA among strains. Alternatively, there may be additional factors essential for methicillin resistance that were not examined in this study.
Conclusions
Both the number and the specific combinations of virulence- and antibiotic resistance-associated genes in S. aureus isolates from the coastal waters of Oahu were found to vary widely. This suggests that more accurate assessments of health risks to bathers may require not only total colony-forming units of S. aureus, but also an assessment of the virulence gene content of those colonies. In addition, the detection of both mecA and femA in methicillin-sensitive isolates suggests that presence of these two genes cannot be used alone as diagnostic indicators for methicillin resistance. Further work to determine accurate molecular indicators of antibiotic resistance will be useful for rapid non-cultivation based diagnostic assays.
Acknowledgements
The authors gratefully acknowledge Dr. M. L. Cooney for his mentoring and leadership in the Marine Science Undergraduate Research Fellowship summer program at the University of Hawaii. Support for the program and for S. Stotts was provided by NSF grant no. 023600 and the University of Hawaii Sea Grant College Program. Additional support for this research was provided by grants from NSF (OCE04-32479) and NIEHS (1P50EF012740-01) through the Pacific Research Center for Marine Biomedicine at the University of Hawaii at Manoa.
References
Baba T, et al. (2002). Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 359, 1819-27.
Betley MJ and JJ Mekalanos. (1985). Staphylococcal enterotoxin A is encoded by phage. Science 229, 185-7.
Boyd EF and H Brüssow. (2002). Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. TRENDS in Microbiology. 10, 521-9.
Chambers HF. (2001). The changing epidemiology of Staphylococcus aureus? Emerging Infectious Diseases. 7, 178-82.
Charoenca N and RS Fujioka. (1993). Assessment of staphylococcus bacteria in Hawaii marine recreational waters. Water Science and Technology. 27, 283-9.
Charoenca N and RS Fujioka. (1995). Association of staphylococcal skin infections and swimming. Water Science and Technology. 31, 11-7.
Diep BA, et al. (2004). Widespread skin and soft-tissue infections due to two methicillin-resistant Staphylococcus aureus strains harboring the genes for Panton-Valentine leucocidin. J Clinical Microbiology. 42, 2080-4.
Fitzgerald JR, et al. (2001). Evolutionary genomics of Staphylococcus aureus: Insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. PNAS. 98, 8821-6.
Gabutti G, et al. (2000). Comparative survival of fecal and human contaminants and use of Staphylococcus aureus as an effective indicator of human pollution. Marine Pollution Bulletin. 40, 697-700.
Kluytmans J, et al. (1997). Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clinical Microbiology Reviews. 10, 505-20.
Kuhl S, et al. (1978). Chromosomal map location of the methicillin-resistant determinant in Staphylococcus aureus. J Bacteriology. 135, 460-5.
Maidhof H, et al. (1991). femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Bacteriology. 173, 3507-13.
Monk AB, et al. (2004). Genetic analysis of Staphylococcus aureus from intravenous drug user lesions. J Medical Microbiology. 53, 223-7.
Moore PCL and JA Lindsay, (2001). Genetic variation among hospital isolates of methicillin-sensitive Staphylococcus aureus: Evidence for horizontal transfer of virulence genes. J Clinical Microbiology. 39, 2760-7.
Nashev D, et al. (2004). Distribution of virulence genes of Staphylococcus aureus isolated from stable nasal carriers. FEMS Microbiology Letters. 233, 45-52.
Peacock SJ, et al. (2002). Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infection and Immunity. 70:4987-96.
Ruzin A, et al. (2001). Molecular genetics of SaPI1 , a mobile pathogemicity island in Staphylococcus aureus. Molecular Microbiology. 41:365-77.
Tillet D and BA Neilan. (2000). Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J Phycology. 36:251-8.
Wilson BA and AA Salyers. (2003). Is the evolution of bacterial pathogens an out of body experience? TRENDS in Microbiology. 11:347-50.